R Lab for Beginners
So, you have a .csv of data, and you want to do some scripted analysis on it in R. Why? Because you think that your analysis will need to be repeated, either by your team (more data will come in) or by other researchers (who may want to validate or reproduce your findings). You don’t want to have to do a multi-step process in Excel or a commercial statistical software product every time, you’d rather have a script you can just run every time you get more data.
This lab takes you through the steps of doing this. If you do the step-by-step version (recommended, if you’re a first-timer), expect to spend an hour on this. If you prefer to just download the final code, you can do this in just a few minutes and simply review the parts that aren’t familiar.
What will we do? We’ll obtain data, bring it into R, look at it, make a few corrections to badly coded data, correct column names, do some summary stats, and visualize the data. We’re not going to get to the point of creating models or doing statistical tests just yet.
Want to watch a video of me doing what I describe below? You can do that here! It’s quite lengthy (40 minutes) but it’s intended for the brand-new user of R and RStudio.
Getting R
You’ll want to (if you haven’t already) install both R and RStudio (you want the free RStudio Desktop version). A little vocab: R is a language, and RStudio is an “IDE” or Integrated Development Environment that makes programming with R much easier. Do you have to use RStudio? No, but it’s highly, highly recommended.
Data Source
In our case, we’re going to use a dataset shared on the UC Irvine machine learning data repository. Specifically, we’re going to use a cervical cancer risk dataset.
Start R Studio
Open RStudio. If it’s your first time ever using RStudio, you might want to watch this brief (watch it muted, there are no words) demo video done by the makers of RStudio.
While you can work in the console (defaults to the lower left pane) to do one-off commands for things you don’t care to save in a script, we’re going to do most of our work in the source pane.
You probably don’t have the source pane visible, unless you’re already working on something there. How do we make it appear? Choose “File”, “New File”, “R Script”. Now you should have an “Untitled” script in the Source pane (defaults to upper left).
Get the Data
We want to get the data into our R environment so we can do things with it. In your source pane (the one that says “Untitled”), type the following:
# Get data
cervical_cancer_data <- read.csv("http://archive.ics.uci.edu/ml/machine-learning-databases/00383/risk_factors_cervical_cancer.csv")The first line is a comment (it won’t be read by R), and the second is an assignment. It will take the csv at the given URL and make that into an object with the name “cervical_cancer_data”.
To run the code, make sure the cursor is in either line 1 or 2 (not below line 2 as if you were typing more), and hit the “Run” button:
You should see an object appear in the Environment pane (upper right by default). You can expand this object (click the triangle) to see more details.
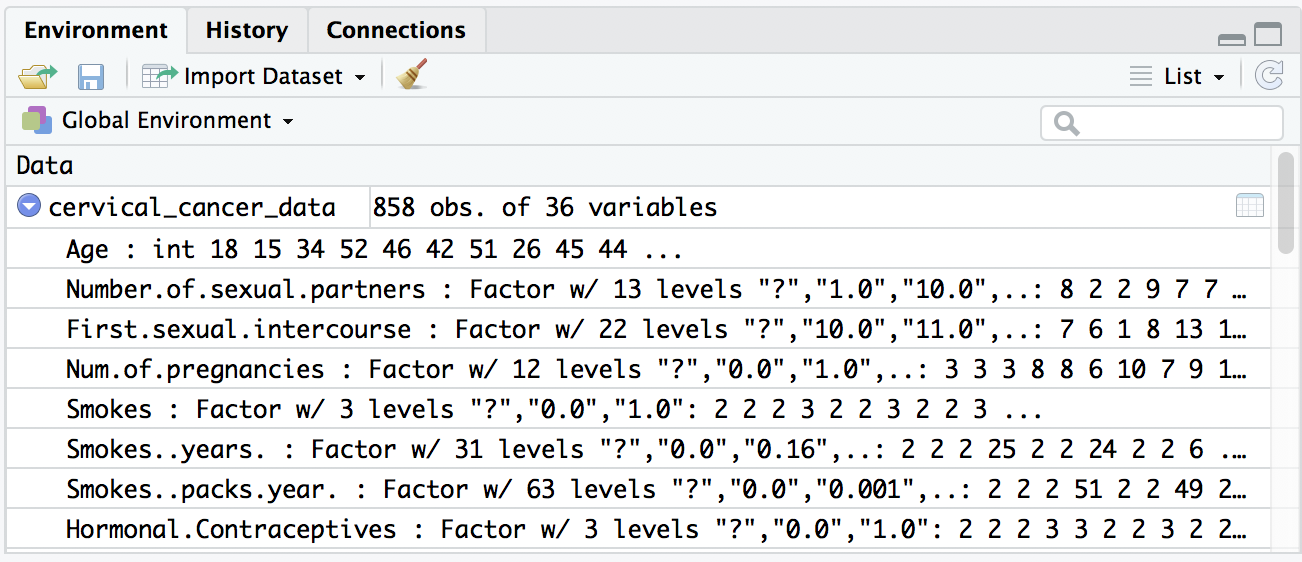
Inspect the data
Uh-oh, we see a few question marks listed in our quick preview of the data. What is that about? Because cervical_cancer_data is a type of data called a data frame, which is like a spreadsheet, you can click on the name of this object in the Environment pane to take a look at the data more closely.
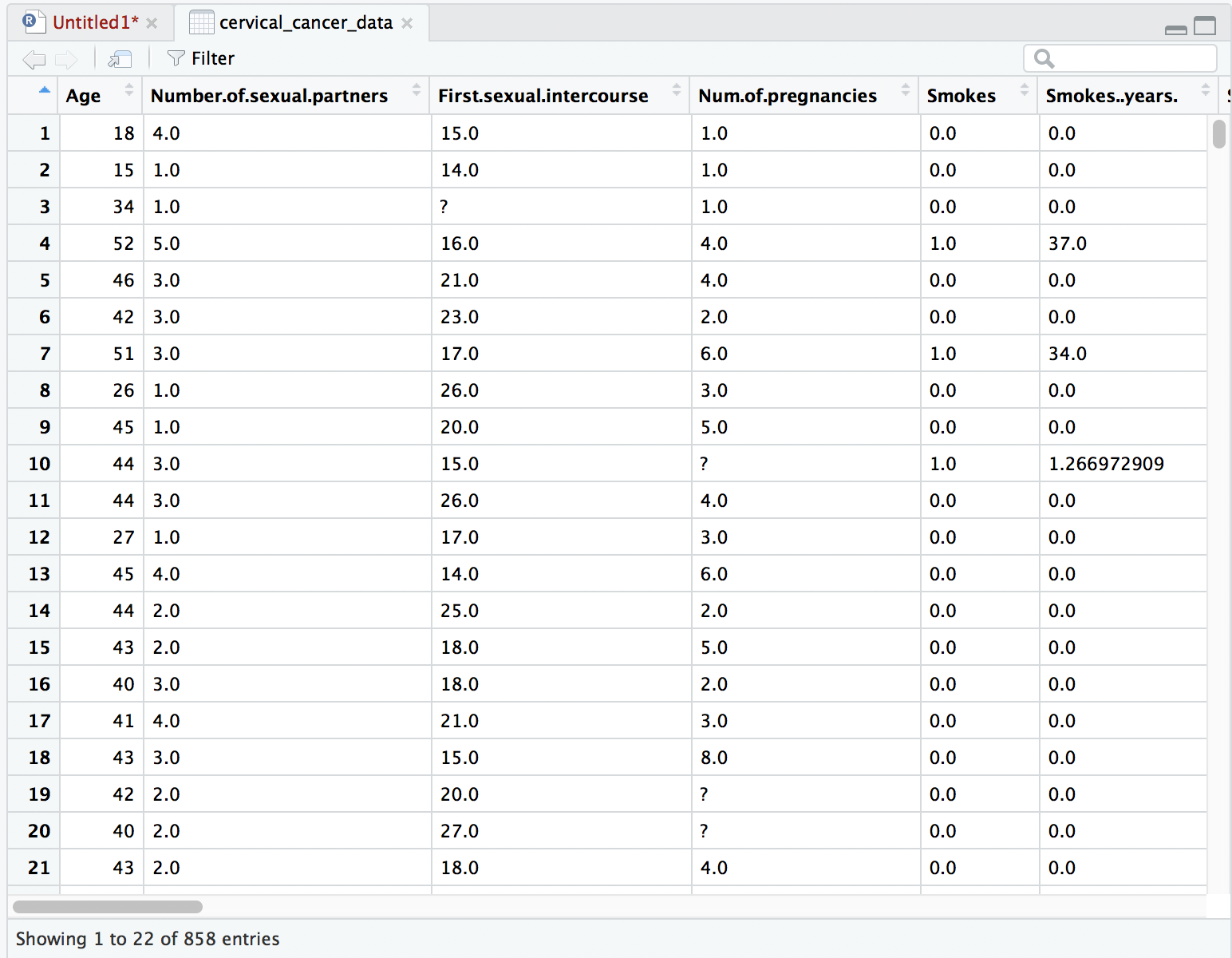
When we look at the data it seems clear that the question marks are intended to be placeholders for “no data available”. We can’t do math on question marks, so that’s going to mess us up. Let’s fix our read.csv command so that it automatically turns question marks into empty values (NA’s). Go back to your source pane and amend the read.csv statement so that it looks like this:
cervical_cancer_data <- read.csv("http://archive.ics.uci.edu/ml/machine-learning-databases/00383/risk_factors_cervical_cancer.csv",
na.strings = c("?"))What we’re doing here is passing in a concatenation (thus the c()) of all the values we want to turn into NA (empty, null) values.
Run that and you’ll see a much nicer quick view in the environment pane. Whereas previously, our data seemed to be all “factor” variables (categorical data), they now look like numeric values:
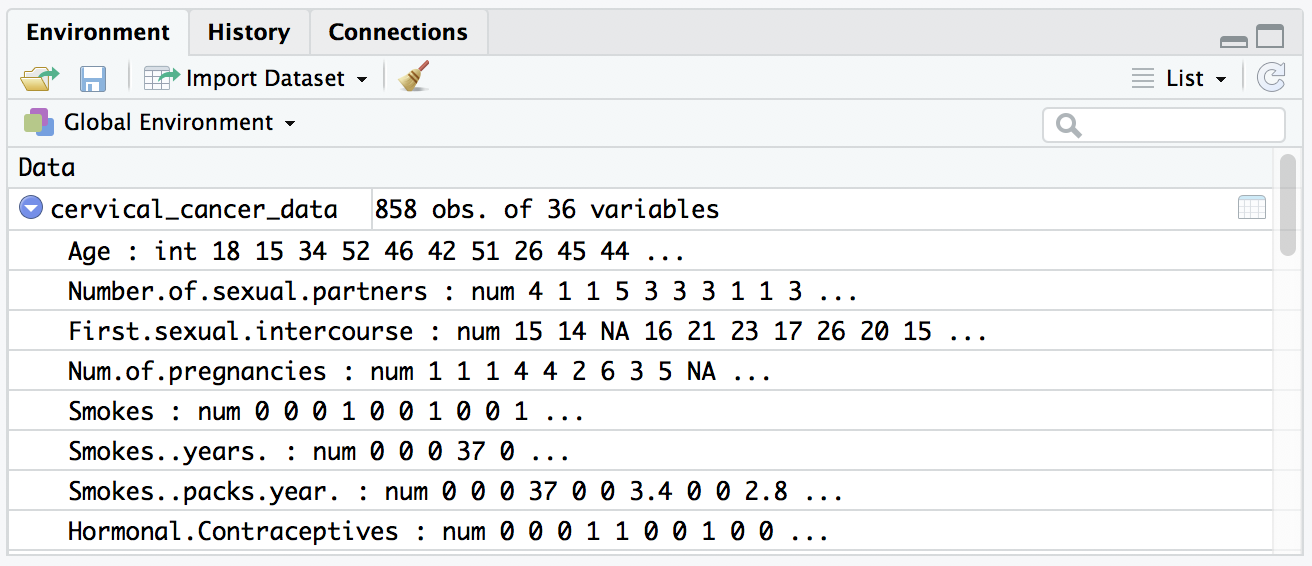
You can also look at the data viewer in your source pane (the tab marked “cervical_cancer_data”), and confirm that the question marks are gone and replaced with NAs.
Statistical Exploration
Let’s do some statistical exploration. Here, we’ll do this in the console (lower left pane). In the console, type summary(cervical_cancer_data) and hit enter. You’ll want a bit more space to view the results, so move your cursor to the area between the upper left and lower left panes until the cursor is a four-way error, then click and drag to resize.
Scroll up and down through the output. Do you notice anything nonsensical?
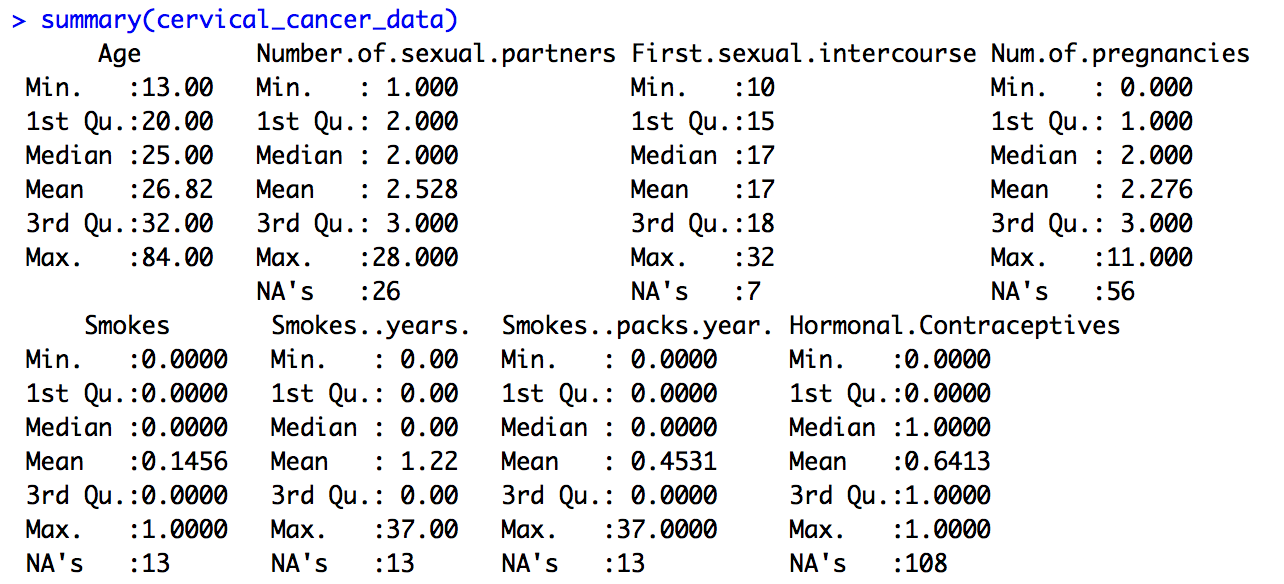
Some things make perfect sense, like mean and median Age and the number of NAs (missing data) in Number.of.sexual.partners. Other things don’t make any sense. For example, Smokes seems to be a category, but it’s being treated like a number, because that’s how it was coded. Finding the third quartile of a category variable doesn’t make sense.
This happens a lot in data, and if you’re not careful you can do something silly like calculate median ZIP code, if you don’t recognize that sometimes numbers aren’t really numerical data, but rather, categorical data. How can we tip off R that variables like these aren’t true numerical data?
We can tell R that these are “factor” variables (where there are one or more categories, like “Male” and “Female”) or “logical” variables (where there are True / False values).
Variable Type
To grasp more fully which variables should be continuous numerical variables and which are categorical or logical, we should check out the data dictionary for this data, which is available (but sparse!) at https://archive.ics.uci.edu/ml/datasets/Cervical+cancer+%28Risk+Factors%29#, under the “Attribute Information”. In this listing, there are two kinds of variables – int, or integer, and bool, or boolean (true/false). In boolean algebra, true is generally coded as 1 and false as 0, so we’ll assume that’s the case here. It’s too bad there’s not a fuller data dictionary with more details!
Also, some of the “bool” values here look like they should be “int” – like smoking pack years. When sharing data, it’s so important to make sure that your coding and data descriptions are complete and accurate. Still, in the un-ideal world of real data, this kind of thing really does crop up, so we’ll take this as a learning opportunity.
Before turning things into factor variables, let’s double-check whether each variable has just two values, 0 and 1, or have more values, indicating that it’s a true integer measure.
We can do that using table(). This will also allow us to check our assumption that True = 1 and False = 0. We can anticipate, for example, that for many columns, like AIDS and Cancer diagnoses, that there will be far more False values than True. If our numerical codes line up as we expect, we’ll see lots more 0s than 1s.
True/False Values
Let’s test using a handful of columns. We can type this into the console, instead of into the source pane. Start by typing table(cervical_cancer_data$STDs.AIDS). All zeros! What about table(cervical_cancer_data$Dx.Cancer)? Mostly zeros, and a handful of ones. Looks like our True/False assumption holds.
In case it’s not obvious, we just used the dollar sign to indicate which column we wanted from our table. There are other ways to indicate which column(s) and/or row(s) we want, but the $ symbol is pretty frequently used when you’re doing analysis across all values of a given variable.
Numeric, or Logical?
Most of the columns seem to make sense, when I compare the data itself (looking just at the data viewer) with the description of the data as int or bool. But there are two smoking-related columns that seem iffy. Let’s double check them with table(). Try table(cervical_cancer_data$Smokes..years.) and table(cervical_cancer_data$Smokes..packs.year.).
Both show a variety of values, and not just “integer” data, but continuous or decimal data. These are clearly not boolean variable types, regardless of what the scanty data dictionary may say.
Renaming Columns
Before we work on setting the variables that need to be recoded as logical or factor variables, let’s rename them. The csv that this data comes from had things like spaces and parentheses, which are rendered in R as dots. This makes the column names a bit clunky.
Let’s get the current names of our variables by typing the following into the console:
names(cervical_cancer_data)
We could, if we wanted to, create our own names, making a concatenation of 36 new names and assigning it to names(cervical_cancer_data), but really all I want to do is make everything lower case and replace all instances of one or more dots with an underscore.
To do this, I’ll first make everything lower case. I’ll add the following to my script, and run the line of code:
names(cervical_cancer_data) <- tolower(names(cervical_cancer_data))Now, to replace the dots with underscores, I’ll use a package called “stringr”. To bring it into my R environment, I’ll add the following to my script and run it:
library(stringr)Note: if you get an error saying “there is no package…”, you’ll need to install the stringr package using the command install.packages("stringr"). You only have to do this once!
Now I can use the stringr package’s command str_replace to find a pattern of one or more (that’s the +) dot characters (that’s the \\.) in the names and replace them with an underscore. The command below uses what’s called a “regular expression”, or regex. It goes beyond the scope of this article, but it’s very very useful to learn. At any rate, add this command to your script and run it.
names(cervical_cancer_data) <- str_replace_all(names(cervical_cancer_data),
"\\.+", "_")Let’s look at the names now. Run names(cervical_cancer_data) in the console. Much better, right? But there are a few that have an underscore at the end of the name. Let’s fix that with another regular expression replacement. Add and execute the following in your script:
names(cervical_cancer_data) <- str_replace_all(names(cervical_cancer_data),
"_$", "")Setting Variable Type
Ok, finally we are ready to set our categorical variables to actually be the right type! We know that our current values are the numbers 0 and 1, where 0 means False and 1 means True. Because this is a special kind of category, we’ll use as.logical() to tell R that it should turn the 0s and 1s into Falses and Trues. Because 0 is conventionally False and 1 is True, we really only have to issue one simple command to turn a column into a True/False variable. Add the following to your script and execute it. Then take a look at your data!
cervical_cancer_data$smokes <- as.logical(cervical_cancer_data$smokes)Now, the annoying thing is that we have to do this for multiple columns. Luckily, R has a way for you to repeat the same command across multiple columns, using the apply() set of functions. This means that instead of copying and pasting the command above for the dozen or so columns that need it, we can do the following. Replace the as.logical() command you just added with the following, and execute it:
# Set True/False columns to be logical
logical_cols <- c("smokes", "hormonal_contraceptives", "iud", "stds",
"stds_condylomatosis", "stds_cervical_condylomatosis",
"stds_vaginal_condylomatosis",
"stds_vulvo_perineal_condylomatosis",
"stds_syphilis", "stds_pelvic_inflammatory_disease",
"stds_genital_herpes", "stds_molluscum_contagiosum",
"stds_aids", "stds_hiv", "stds_hepatitis_b", "stds_hpv",
"dx_cancer", "dx_cin", "dx_hpv", "dx", "hinselmann",
"schiller", "citology", "biopsy")
cervical_cancer_data[logical_cols] <- lapply(cervical_cancer_data[logical_cols], as.logical)Take a look at your data now – it should be much more intuitive, and ready for analysis. Let’s recap. You should have the code listed below. If you want, you can re-run everything to make sure that your data is in synch with this lab. You can highlight the lines of code you want to run in your script and hit “run” to execute multiple lines, or just put your cursor in the top line and hit run a few times to step through the code line-by-line.
# Get data
cervical_cancer_data <- read.csv("http://archive.ics.uci.edu/ml/machine-learning-databases/00383/risk_factors_cervical_cancer.csv",
na.strings = c("?"))
# Make variable names better
names(cervical_cancer_data) <- tolower(names(cervical_cancer_data))
library(stringr)
names(cervical_cancer_data) <- str_replace_all(names(cervical_cancer_data),
"\\.+", "_")
names(cervical_cancer_data) <- str_replace_all(names(cervical_cancer_data),
"_$", "")
# Set True/False columns to be logical
logical_cols <- c("smokes", "hormonal_contraceptives", "iud", "stds",
"stds_condylomatosis", "stds_cervical_condylomatosis",
"stds_vaginal_condylomatosis",
"stds_vulvo_perineal_condylomatosis",
"stds_syphilis", "stds_pelvic_inflammatory_disease",
"stds_genital_herpes", "stds_molluscum_contagiosum",
"stds_aids", "stds_hiv", "stds_hepatitis_b", "stds_hpv",
"dx_cancer", "dx_cin", "dx_hpv", "dx", "hinselmann",
"schiller", "citology", "biopsy")
cervical_cancer_data[logical_cols] <- lapply(cervical_cancer_data[logical_cols], as.logical)Graphical Exploration
Let’s see what the distributions of our data are. We can use cross-tabulations and histograms to take quick peeks.
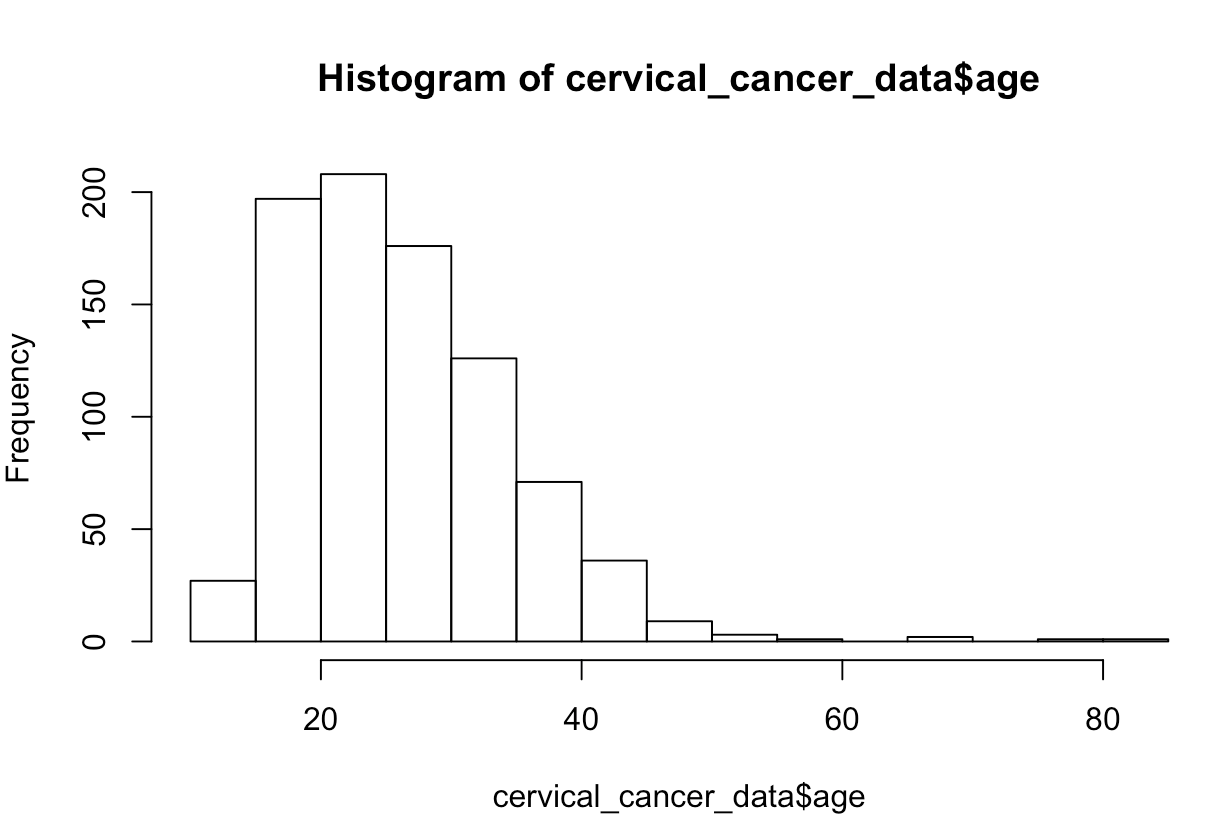
What if we want to see the distribution of ages? Try this:
hist(cervical_cancer_data$age)In your plot window (defaults to lower right), you should see an image like this:
What about a boxplot of number of sexual partners?
boxplot(cervical_cancer_data$number_of_sexual_partners)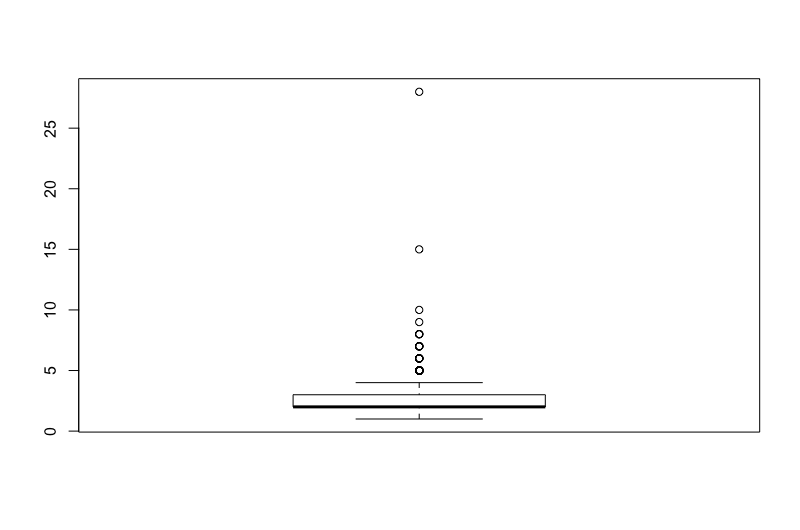
Want to see if there’s a relationship between years of smoking and number of STDs?
plot(x=cervical_cancer_data$smokes_years, y=cervical_cancer_data$stds_number)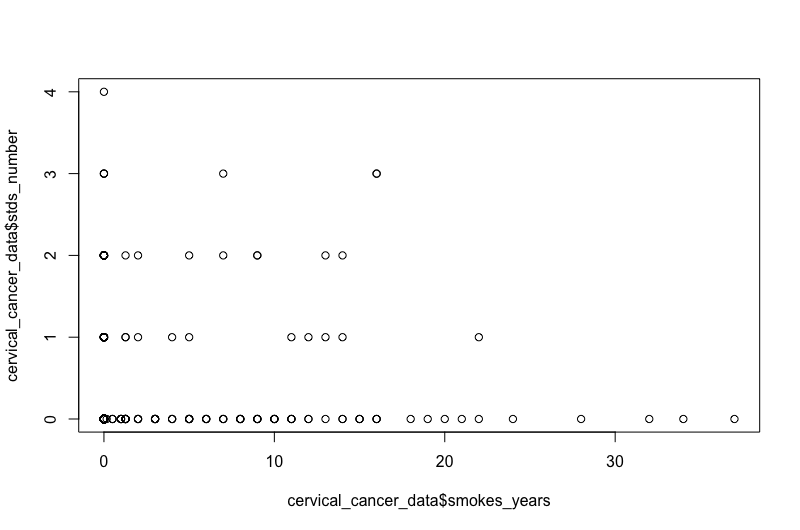
For these and most, if not all, R commands, if you need help you can type ?plot() or ?boxplot(), etc.
Obviously these graphics aren’t publication-ready, but are helpful for understanding the data and coming up with preliminary answers to questions like:
- Is my data normally distributed?
- Does there seem to be a relationship?
- Are there any weird outliers?
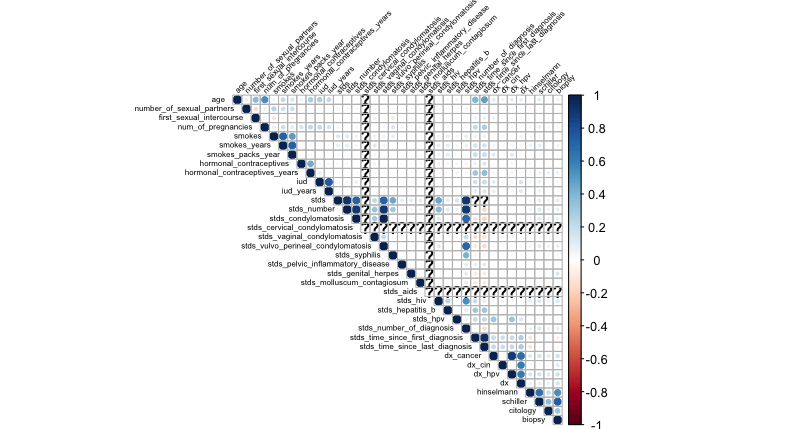
What if we want to create a correlation matrix heatmap to see how different factors correlate?
Give this a shot in your console. Remember that if you get an error on the library() command, you can usually solve that by issuing, just once, install.packages("name_of_package") for the package that failed on library(). In these commands, we’ll create a matrix that has numerical correlation measurements, then plot the upper triangle of the matrix with the title text in black, tilted at 45 degrees, and at half the typical size.
# Create and display a correlation matrix
corr_matrix <- cor(cervical_cancer_data, use="pairwise.complete.obs")
library(corrplot)
corrplot(corr_matrix, type = "upper", tl.col = "black", tl.srt = 45, tl.cex = .5)Hopefully you see a nice correlation heatmap like this one:
Non-Graphical Exploration
Issue the following in your console, which gives you a table of genital herpes and cancer diagnoses:
xtabs(~ dx_cancer + stds_genital_herpes, data=cervical_cancer_data)Or, try this one, which gives you much the same appearance. Note that I give the dimension names explicitly here.
table(cervical_cancer_data$stds_genital_herpes, cervical_cancer_data$dx_cancer, dnn=c("HSV", "Cancer"))In R, there are frequently many ways to get the same information. You will start to develop your own style and preferred commands, so don’t worry if you see people using different code than what you’re accustomed to.
You’ve already seen how to use summary() to get basic summary stats on your data. This time, run it to identify which columns might not be good variables to use in the research because of lots of missing data.
summary(cervical_cancer_data)If you look at the bottom of each variable’s summary stats, you can see the number of NA’s. There are rather a few variables that are missing lots of values. This includes most of the STD fields. We might want to clean up our data by removing these columns (runs the risk of abandoning a great explanatory variable), or remove the rows for which data is absent (runs the risk of subject selection bias), or by interpolating data, perhaps by putting in a median value (probably not valid here).
Let’s leave things there, and in a second lab, we’ll work on basic inferential statistics!
