A/B testing 1 is a statistical way of comparing two (or more) techniques, typically the current approach against a new option. A/B testing aims to determine not only which technique performs better but also to understand whether the difference is statistically significant. A/B testing usually considers only two techniques using one measurement, but it can be applied to any finite number of techniques and measures.
Accuracy 1: The fraction of predictions that a classification model got right. In multi-class classification, accuracy is defined as follows:
In binary classification, accuracy has the following definition:
In medicine, we often deal with unbalanced classes, such as in the case of rare diseases. In a case like this, Balanced Accuracy may prove to be a better measure.
It may also be useful to ignore traditional accuracy measures and instead judge a model’s effectiveness using Positive Predictive Value (PPV) (also known as precision), sensitivity (also known as recall), specificity, or some other measure that tunes your model to the correct type of optimization for the problem at hand.
Algorithm 2: a process or set of rules to be followed in calculations or other problem-solving operations.
Anaconda : a Python distribution that includes just about everything you need to begin using Python for data analysis. It’s freely available at the Anaconda distribution website.
Application programming interface (API) : programmatic communication between different software components, allowing information to flow between two different systems (say, REDCap and an R script) as needed, without human interaction.
Artificial intelligence 2: the power of a machine to copy intelligent human behavior. At times this term gets conflated with machine learning.
Attribute 1: Synonym for feature. Attributes may also refer to characteristics pertaining to individuals.
AUC (Area under the ROC Curve) 1: An evaluation metric that considers all possible classification thresholds.
The Area Under the ROC curve is the probability that a classifier will be more confident that a randomly chosen positive example is actually positive than that a randomly chosen negative example is positive.
Back Propagation or Backpropagation1: The primary algorithm for performing gradient descent on neural networks. First, the output values of each node are calculated (and cached) in a forward pass. Then, the partial derivative of the error with respect to each parameter is calculated in a backward pass through the graph.
Bag of Words1: A representation of the words in a phrase or passage, irrespective of order. For example, bag of words represents the following three phrases identically:
- the dog jumps
- jumps the dog
- dog jumps the
Each word is mapped to an index in a sparse vector, where the vector has an index for every word in the vocabulary. For example, the phrase the dog jumps is mapped into a feature vector with non-zero values at the three indices corresponding to the words the, dog, and jumps. The non-zero value can be any of the following:
- A 1 to indicate the presence of a word.
- A count of the number of times a word appears in the bag. For example, if the phrase were the maroon dog is a dog with maroon fur, then both maroon and dog would be represented as 2, while the other words would be represented as 1.
- Some other value, such as the logarithm of the count of the number of times a word appears in the bag.
Batch1: The set of examples used in one iteration (that is, one gradient update) of model training.
Batch Size1: The number of examples in a batch. For example, the batch size of SGD is 1, while the batch size of a mini-batch is usually between 10 and 1000. Batch size is usually fixed during training and inference; however, TensorFlow does permit dynamic batch sizes.
Bayesian Neural Network1 A probabilistic neural network that accounts for uncertainty in weights and outputs. A standard neural network regression model typically predicts a scalar value; for example, a model predicts a house price of 853,000. By contrast, a Bayesian neural network predicts a distribution of values; for example, a model predicts a house price of 853,000 with a standard deviation of 67,200. A Bayesian neural network relies on Bayes’ Theorem to calculate uncertainties in weights and predictions. A Bayesian neural network can be useful when it is important to quantify uncertainty, such as in models related to pharmaceuticals. Bayesian neural networks can also help prevent overfitting.
Bias1 can refer to several ideas, and we treat these terms as three different glossary entries.
- bias as an ethical term (Ethical Bias), referring to unfair or inaccurate representation of reality due to prejudice or systematic error.
- bias as a mathematical term (Bias Term) in machine learning, also known as an intercept.
- bias as a measure of the difference between predictions and actual values in modeling (prediction bias).
Bias Term1: An intercept or offset from an origin. Bias (also known as the bias term) is referred to as b or w0 in machine learning models. For example, bias is the b in the following formula:
Not to be confused with ethical bias or prediction bias.
Bigram1: in NLP, an N-gram in which N=2. In other words, a phrase of word length two, such as:
- a phrase
- phrase of
- of word
- word length
- length two
Binary Classification1: A type of classification task that outputs one of two mutually exclusive classes. For example, a machine learning model that evaluates email messages and outputs either “spam” or “not spam” is a binary classifier.
Binning (or “Bucketing”)1: Converting a (usually continuous) feature into multiple binary features called buckets or bins, typically based on value range. For example, instead of representing temperature as a single continuous floating-point feature, you could chop ranges of temperatures into discrete bins. Given temperature data sensitive to a tenth of a degree, all temperatures between 0.0 and 15.0 degrees could be put into one bin, 15.1 to 30.0 degrees could be a second bin, and 30.1 to 50.0 degrees could be a third bin.
Blocking or Block design – Controlling variability in an outcome by creating groups based on characteristics you can’t control, like sex.
Boosting1: A machine learning technique that iteratively combines a set of simple and not very accurate classifiers (referred to as “weak” classifiers) into a classifier with high accuracy (a “strong” classifier) by upweighting the examples that the model is currently misclassfying.
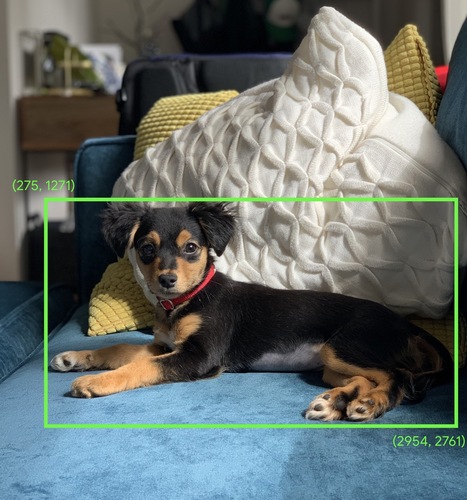
Bounding Box1: In an image, the (x, y) coordinates of a rectangle around an area of interest, such as the dog in the image below.
Centroid1 The center of a cluster as determined by a k-means or k-median algorithm. For instance, if k is 3, then the k-means or k-median algorithm finds 3 centroids.
Cloud computing : an off-site (“somewhere in the cloud of the internet”) shared system of resources (like servers, applications, storage) usable by many individuals and groups. Common cloud computing providers include Amazon Web Services (AWS), Google Cloud Platform (GCP), Rackspace, Microsoft Azure, etc. Cloud computing can be cost effective and robust to changes in needs (say, if your application sometimes spikes in its resource needs).
Common data element (CDE) 2: piece of data common to multiple datasets across different studies (may be universal or domain-specific).
Crowdsourcing 2: a distributed model in which individuals or groups obtain services, ideas, or content from a large, relatively open and often rapidly-evolving group of internet users.
Database/Data repository 2: Data storage that stores, organizes, validates, and makes accessible core data related to a particular system or systems. Generally speaking, most databases and data repositories rely on a “relational” database model (which use various flavors of SQL), which has interrelated tables, each of which has rows and columns. There are other types of databases which do not have this kind of tabular format. While very different from each other, these non-relational, non-SQL database types are often grouped under the same term, noSQL.
DataCamp is a data education website with free and paid courses on R, Python, and SQL. Benefits of using DataCamp include a browser-only interface (nothing to download) and structured courses that walk you through the basics, step by step. If you’ve never used R, Python, or SQL, it might be worth trying one of their free intro classes to get you over the initial inertia of not knowing how to start!
Data commons 2: a shared virtual space in which scientists can work with the digital objects of biomedical research such as data and analytical tools.
Data ecosystem 2: a distributed, adaptive, open system with properties of self-organization, scalability, and sustainability inspired by natural ecosystems.
Data integrity 2: the accuracy and consistency of data stored in a database/data repository, data warehouse, data mart or other construct.
Data science 2: interdisciplinary field of inquiry in which quantitative and analytical approaches, processes, and systems are developed and used to extract knowledge and insights from increasingly large and/or complex sets of data.
Dataset 2: collection of related sets of information composed of separate elements that can be manipulated computationally as a unit.
Data visualization 2: effort to help people understand the significance of data by placing it in a visual context. Patterns, trends and correlations that might go undetected can be exposed and recognized more easily with data-visualization techniques.
Deep learning 2: type of machine learning in which each successive layer uses output from the previous layer as input; similar to communication patterns in a biological nervous system.
Domain-specific 2: for biomedical data, designed and intended for use in studies of a particular topic, disease or condition, or body system (compare to universal).
Effect Size : a standardized measure of how large the difference detected by a statistical test is. It’s a proportion of the difference discovered over the dispersion of the samples. To make this more concrete, imagine discovering a statistically significant difference in sleep amounts between insomniac patients taking a placebo and insomniac patients taking a new sleep medication. You could have a statistically significant difference at p = .03, where the difference in means is 12 minutes, or a statistically significant difference at p = .03, where the differnce in mean is 2 hours. Clearly, one finding is more interesting. Effect size allows us to go beyond statistical significance and get some idea of how important the discovered difference is. It is important to realize that it is easier to detect a large effect size than a smaller one, and that knowing the effect size you’re interested in will affect your power and sample size.
Elastic describes a service that can get bigger as needed. This is an advantage of cloud computing platforms, which can be set up to use more servers (or other resources) as needed. For example, a website might be set up to add servers when traffic exceeds 70% of maximum, so that no one gets an “unable to connect” error when going to the website.
Electronic medical record (EMR) or Electronic health record (EHR) 2: digital version of a patient’s paper chart. EHRs are real-time, patient- centered records that make information available instantly and securely to authorized users. At CHOP, our EHR system is Epic.
Enterprise system 2: computer hardware and software used to satisfy the needs of an organization rather than individual users. “Enterprise-grade” usually signifies high capacity, customer support, robust applications that support business functions that cannot fail.
Ethical bias1
- Stereotyping, prejudice or favoritism towards some things, people, or groups over others. These biases can affect collection and interpretation of data, the design of a system, and how users interact with a system. Forms of this type of bias include:
- automation bias
- confirmation bias
- experimenter’s bias
- group attribution bias
- implicit bias
- in-group bias
- out-group homogeneity bias
- Systematic error introduced by a sampling or reporting procedure. Forms of this type of bias include:
- coverage bias
- non-response bias
- participation bias
- reporting bias
- sampling bias
- selection bias
Not to be confused with the bias term in machine learning models or prediction bias.
Exascale computing 2: a computer system capable performing one quintillion (10^18) calculations per second.
Excel is spreadsheet software that many people use for research. This is a bad idea for multiple reasons: it’s not reproducible (no syntax file or scripted play-back available of the manipulations you do, unless you use VBA), it makes assumptions about data that are often wrong (for example, genomics research is rife with gene name errors caused by Excel), and it’s not rigorous with data (for example, you can accidentally sort just one column instead of a whole sheet and end up with erroneous data). If you typically use Excel to work with data, consider learning R.
Extramural 2: research or other activities supported by NIH and conducted by external organizations, and funded by grants, contracts, or cooperative agreements from NIH.
Genotype 2: the genetic make-up of an individual organism
git is a version control program that allows you to track changes to your code (or other text-based files), understand who changed a file and why, manage multiple canonical versions (say, if you have one version for use with PHI and one with de-identified data), and go back to older versions as required.
GitHub is a website that uses git and provides a nice user interface for working with git. GitHub also offers a useful graphical software package called GitHub Desktop, which is useful for working with git without having to know how to use the command line. It’s great for git beginners.
Gradient Descent is an optimization technique used in machine learning to find the right weights to use with features to make predictions. It relies on principles of multivariable calculus to determine which direction weights should change in, and how much, to improve the accuracy of prediction.
Hardening 2: process of optimizing a tool or algorithm to industry standards to ensure efficiency, ease of use, security, and utility.
Hardware 2: collection of physical parts of a computer system.
Indexing 2: methods to allow data finding and retrieving.
Interoperability 2: in computer systems, the ability to exchange and make use of information from various sources and of different types.
Intramural 2: research or other activities conducted by, or in support of, NIH employees on its Bethesda, Maryland, campus or at one of the other NIH satellite campuses across the country.
Jupyter Notebook (formerly iPython): This is a literate statistical programming approach for Python. A Jupyter Notebook is a “REPL” (Read, Execute, Print Loop) which allows you to work interactively with Python, executing code bit by bit, changing it up, and adding markdown. This means you don’t have to write and execute a whole program to check that your code works. This is great for exploratory work as well as for showing output for other users of your code.
Knowledgebase 2: virtual resource that accumulates, organizes, and links growing bodies of information related to core datasets
Machine Learning is a broad category of heavily computational analysis and prediction methods that create models of data which depend on finding patterns that organize data (e.g. unsupervised learning like cluster analysis and feature detection and extraction) and predict outcomes (e.g. supervised learning like classification or regression predictions). Machine learning can relatively simple and be very reminiscent of traditional mathematical approaches like linear regression, or quite complex, as when multiple models are combined (“ensembled”) to improve predictive performance. Machine learning can result in surprisingly accurate predictions, but it can also admit of significant problems, like models that are not easily actionable, or models that are too narrowly focused on a small set of data (overfitting).
Metadata 2: data that describe other data. Examples include title, abstract, author, and keywords (publications); organization and relationships of digital materials; and file types or modification dates.
Multiple Factor or Multifactorial: Describes an experiment or observation in which an outcome (or dependent) variable is measured in relationship to more than one explanatory (or independent) variable. For example, the study of adult BMI as a function of birth weight and Adverse Childhood Events (ACE) score.
NIH IC 2: NIH Institute or Center
*omics 2: collective characterization and measurement of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms. Examples include genomics, proteomics, metabolomics, and others.
Petascale computing 2: a computer system capable performing one quadrillion (10^15) calculations per second. It is currently used in weather and climate simulation, nuclear simulations, cosmology, quantum chemistry, lower-level organism brain simulation, and fusion science.
Phenotype 2: the set of observable characteristics of an individual resulting from the interaction of its genotype with the environment.
Platform 2: group of technologies (software and hardware) upon which other applications, processes, or technologies are developed.
Platform as a Service (PaaS) 2: a type of cloud computing that allows users to develop, run, and manage applications without the complexity of building and maintaining an overarching infrastructure
Power: The probability that a statistical test will correctly reject the null hypothesis when the alternative hypothesis is true. Power is the ability to avoid false negatives, and is closely related to effect size and sample size. Often, we choose a .8, or 80% power, which means that we expect our test to be able to detect true positives (where the alternative hypothesis is true) 80% of the time.
Provenance 2: timeline of ownership, location, and modification.
Python is a programming language that has multiple uses. It can build desktop applications, websites, perform image manipulation and analyses, and do advanced statistical and data-centric workflows. Unlike R, python is multi-purpose. Like R, it’s a true programming language which supports script-based reproducible research. If you have experience programming in other languages like Java or C++, you might like Python more than R, especially if you are doing natural language processing or web scraping or need to do tasks beyond data analysis, such as building a website or creating an application. On the other hand, if you’ve mostly used Excel or a commercial product like SPSS to do your research and don’t have experience programming, R makes a better starting point.
R is a statistical programming language which is open source – it’s free and anyone can contribute to its development, or create packages that extend its usefulness. It is different than Python because it’s not a multi-purpose language used for all sorts of things – it’s built for statistical analysis. R is also different from commercial statistical software like SAS and SPSS in that it is not a point-and-click solution. R is widely used in academia because it can be tailored to very specialized analyses, supports true reproducible research, is free, and gets innovative analytics support earlier than commercial software. R is best used within the RStudio development environment (also free!).
Reproducibility: There are periodic kerfuffles about whether the right term is reproducibility or replicability, but here we mean an approach to research that makes it easy to re-run an analysis on a new (or updated) dataset, with a minimum of problems. An example of research that’s not reproducible would be making a lot of manual choices in Excel, like cleaning data, removing outliers, and using formulas to create new variables and analyses. This is very difficult to reproduce (as you know, if you’ve ever tried to replicate a finding based on a methods section!). An example of reproducible research would be a sample file that has publicly-sharable data, or fabricated example data, and an R script that takes a file with that format, conducts analyses, and creates statistical output like p values. That would be fairly easy for a different researcher to pick up and work with.
Retirement (of data) 2: the practice of shutting down redundant or obsolete business applications while retaining access to the historical data.
Sample Size : the number of observed entities (in our case, often animal or human subjects) used in a research project. The larger the sample size, the more specific the statistical tests can be. When research aims to find a small effect size , a large sample size is required to maintain the same power.
Software 2: programs and other operating information used by a computer.
Software as a Service (SaaS) 2: software licensing and delivery model in which software is licensed on a subscription basis and is centrally hosted.
Sparse Vector1:
A vector whose values are mostly zeroes. See also sparse feature.
Sparsity (or Missingness)1:
The number of elements set to zero (or null) in a vector or matrix divided by the total number of entries in that vector or matrix. For example, consider a 10x10 matrix in which 98 cells contain zero, null, or NaN, depending on context. The calculation of sparsity is as follows:
Feature sparsity refers to the sparsity of a feature vector; model sparsity refers to the sparsity of the model weights.
Squared Hinge Loss1: The square of the hinge loss. Squared hinge loss penalizes outliers more harshly than regular hinge loss.
Squared Loss1: The loss function used in linear regression. (Also known as L2 Loss.) This function calculates the squares of the difference between a model’s predicted value for a labeled example and the actual value of the label. Due to squaring, this loss function amplifies the influence of bad predictions. That is, squared loss reacts more strongly to outliers than L1 loss.
State1: In reinforcement learning, the parameter values that describe the current configuration of the environment, which the agent uses to choose an action.
State-Action Value Function1: Synonym for Q-function.
Stationarity1: A property of data in a dataset, in which the data distribution stays constant across one or more dimensions. Most commonly, that dimension is time, meaning that data exhibiting stationarity doesn’t change over time. For example, data that exhibits stationarity doesn’t change from September to December.
Step1:
In machine learning, a forward and backward evaluation of one batch.
Step size1:
Synonym for learning rate.
Stochastic Gradient Descent (SGD) is a form of gradient descent that determines the next “step” (next set of weights to try) by looking at a single example from a dataset and estimating the gradient from that sample of 1.
Structural Risk Minimization (SRM)1: An algorithm that balances two goals:
- The desire to build the most predictive model (for example, lowest loss).
- The desire to keep the model as simple as possible (for example, strong regularization).
For example, a function that minimizes loss+regularization on the training set is a structural risk minimization algorithm.
For more information, see http://www.svms.org/srm/.
Contrast with empirical risk minimization.
Supervised Machine Learning1: Training a model from input data and its corresponding labels. Supervised machine learning is analogous to a student learning a subject by studying a set of questions and their corresponding answers. After mastering the mapping between questions and answers, the student can then provide answers to new (never-before-seen) questions on the same topic. Compare with unsupervised machine learning, which does not give labels.
Synthetic Feature 1 In machine learning, a feature not present among the input features, but created from one or more of them. Kinds of synthetic features include:
- Binning / bucketing a continuous feature into range bins.
- Multiplying (or dividing) one feature value by other feature value(s) or by itself.
- Creating a feature cross.
Features created by normalizing or scaling alone are not considered synthetic features.
System integrator/system engineer 2: individual who refines and hardens tools from academia to improve user design, authentication and testing, and optimize productivity, efficiency, and outcomes/performance.
Tabular Q-learning 1: In reinforcement learning, implementing Q-learning by using a table to store the Q-functions for every combination of state and action.
Target 1: Synonym for label, a representation of the actual value of an observation.
Target Network 1: In Deep Q-learning, a neural network that is a stable approximation of the main neural network, where the main neural network implements either a Q-function or a policy. Then, you can train the main network on the Q-values predicted by the target network. Therefore, you prevent the feedback loop that occurs when the main network trains on Q-values predicted by itself. By avoiding this feedback, training stability increases.
Temporal Data 1: Data recorded at different points in time. For example, winter coat sales recorded for each day of the year would be temporal data.
Tensor 1: The primary data structure in TensorFlow programs. Tensors are N-dimensional (where N could be very large) data structures, most commonly scalars, vectors, or matrices. The elements of a Tensor can hold integer, floating-point, or string values.
TensorBoard 1: The dashboard that displays the summaries saved during the execution of one or more TensorFlow programs.
TensorFlow 1: A large-scale, distributed, machine learning platform developed by Google. The term also refers to the base API layer in the TensorFlow stack, which supports general computation on dataflow graphs.
Although TensorFlow is primarily used for machine learning, you may also use TensorFlow for non-ML tasks that require numerical computation using dataflow graphs.
Tensor Processing Unit (TPU) 1: An application-specific integrated circuit (ASIC) that optimizes the performance of machine learning workloads. These ASICs are deployed as multiple TPU chips on a TPU device.
Termination Condition 1: In reinforcement learning, the conditions that determine when an episode ends, such as when the agent reaches a certain state or exceeds a threshold number of state transitions. For example, in tic-tac-toe (also known as noughts and crosses), an episode terminates either when a player marks three consecutive spaces or when all spaces are marked.
Test Set 1: In machine learning, the subset of the dataset used to test a model after the model has gone through initial vetting by the validation set.
Contrast with training set and validation set.
Time Series Analysis 1: A subfield of machine learning and statistics that analyzes temporal data. Many types of machine learning problems require time series analysis, including classification, clustering, forecasting, and anomaly detection. For example, you could use time series analysis to forecast the future sales of winter coats by month based on historical sales data.
Tower 1: A component of a deep neural network that is itself a deep neural network without an output layer. Typically, each tower reads from an independent data source. Towers are independent until their output is combined in a final layer.
Training 1: In machine learning, the process of determining the ideal parameters that make up a model.
Training Set 1: In machine learning, the subset of the dataset used to train a model.
Contrast with validation set and test set.
Transfer Learning1: Transferring information from one machine learning task to another. For example, in multi-task learning, a single model solves multiple tasks, such as a deep model that has different output nodes for different tasks. Transfer learning might involve transferring knowledge from the solution of a simpler task to a more complex one, or involve transferring knowledge from a task where there is more data to one where there is less data.
Most machine learning systems solve a single task. Transfer learning is a baby step towards artificial intelligence in which a single program can solve multiple tasks.
Trigram 1: In NLP, an N-gram in which N=3. In other words, a phrase of word length three, such as:
- a phrase of
- phrase of word
- of word length
- word length three
True Negative (TN) 1 An example in which the model correctly predicted the negative class. For example, the model inferred that a particular email message was not spam, and that email message really was not spam.
True Positive (TP) 1 An example in which the model correctly predicted the positive class. For example, the model inferred that a particular email message was spam, and that email message really was spam.
True Positive Rate (TPR) 1 Synonym for recall or sensitivity. The proportion of positives that a model successfully detected. That is:
True positive rate is represented as the y-axis in an ROC curve.
Unique identifiers 2: an alphanumeric string (such as 1a2b3c) used to uniquely identify an object or entity on the internet.
Universal 2: for biomedical data, usable in research studies regardless of the specific disease or condition of interest (compare to domain-specific).
Producing a model with poor predictive ability because the model hasn’t captured the complexity of the training data. Many problems can cause underfitting, including:
- Training on the wrong set of features.
- Training for too few epochs or at too low a learning rate.
- Training with too high a regularization rate.
- Providing too few hidden layers in a deep neural network.
Unlabeled Example 1 An example that contains features but no label. Unlabeled examples are the input to inference. In semi-supervised and unsupervised learning, unlabeled examples are used during training.
Unsupervised Machine Learning 1
Training a model to find patterns in a dataset, typically an unlabeled dataset.
The most common use of unsupervised machine learning is to cluster data into groups of similar examples. For example, an unsupervised machine learning algorithm can cluster songs together based on various properties of the music. The resulting clusters can become an input to other machine learning algorithms (for example, to a music recommendation service). Clustering can be helpful in domains where true labels are hard to obtain. For example, in domains such as anti-abuse and fraud, clusters can help humans better understand the data.
Another example of unsupervised machine learning is principal component analysis (PCA). For example, applying PCA on a dataset containing the contents of millions of shopping carts might reveal that shopping carts containing lemons frequently also contain antacids.
Compare with supervised machine learning.
Validation 1: A process used, as part of training, to evaluate the quality of a machine learning model using the validation set. Because the validation set is disjoint from the training set, validation helps ensure that the model’s performance generalizes beyond the training set.
Contrast with test set and training set.
Validation Set 1: In machine learning, a subset of the dataset -disjoint from the training set — used in validation.
Contrast with training set and test set.
Version Control : Version control is a system that helps tracks various versions of files (usually code). To grasp the concept, think about “track changes” in Microsoft Word or editing a document Google Docs, where you can see who made what changes and why, and go back to any version you need to. The most popular version control system is git, which is a distributed version control system. Many people use git within GitHub, which offers not only version control, but file hosting and a simple user interface that may be easier for beginners to use than command-line git.
Wearables 2: devices that can be worn by a consumer that collect data to track health.
Weight 1: A coefficient for a feature in a linear model, or an edge in a deep network. The goal of training a linear model is to determine the ideal weight for each feature. If a weight is 0, then its corresponding feature does not contribute to the model.
Wide Model1 A linear model that typically has many sparse input features. We refer to it as “wide” since such a model is a special type of neural network with a large number of inputs that connect directly to the output node. Wide models are often easier to debug and inspect than deep models. Although wide models cannot express nonlinearities through hidden layers, they can use transformations such as feature crossing and bucketization (binning to model nonlinearities in different ways.
Contrast with deep model.
Width1: In the context of neural networks, the number of neurons in a particular layer of the network.
Workflow 2: defined series of tasks for processing data.
-
Definition taken from or adapted from a Google Machine Learning Glossary ↩ ↩2 ↩3 ↩4 ↩5 ↩6 ↩7 ↩8 ↩9 ↩10 ↩11 ↩12 ↩13 ↩14 ↩15 ↩16 ↩17 ↩18 ↩19 ↩20 ↩21 ↩22 ↩23 ↩24 ↩25 ↩26 ↩27 ↩28 ↩29 ↩30 ↩31 ↩32 ↩33 ↩34 ↩35 ↩36 ↩37 ↩38 ↩39 ↩40 ↩41 ↩42 ↩43 ↩44 ↩45 ↩46 ↩47 ↩48 ↩49 ↩50 ↩51 ↩52 ↩53 ↩54 ↩55 ↩56 ↩57
-
Definition taken from or adapted from NIH Strategic Plan for Data Science ↩ ↩2 ↩3 ↩4 ↩5 ↩6 ↩7 ↩8 ↩9 ↩10 ↩11 ↩12 ↩13 ↩14 ↩15 ↩16 ↩17 ↩18 ↩19 ↩20 ↩21 ↩22 ↩23 ↩24 ↩25 ↩26 ↩27 ↩28 ↩29 ↩30 ↩31 ↩32 ↩33 ↩34 ↩35 ↩36 ↩37 ↩38 ↩39 ↩40